Integrated Genomic Breeding
Exploring and utilization of useful traits from diverse rice genetic resources
Rice is cultivated all around the world and shows numerous morphological and physiological differences in the form of phenotypic variations. Some of these variations have been used as genetic resources to improve rice plants so that they better satisfy human needs. Phenotypic variations are considered to be genetically controlled by the collective function of a large number of genes on rice genomes However, the genetic bases and biological functions of most of them are still unknown, which has prevented us from wider practical application of rice germplasms. We exploit useful phenotypic variations from a wide range of rice germplasms and clarify the genes involved with biological functions by aid of recent advances in genome information and technology. Also, we try to develop new breeding materials and propose more effective breeding methodologies.
 |
|
|---|
Development of remote cross breeding in rice via polyploidization
 For breeding of super rice varieties with useful new genes, it is important to promote allelic exchange in existing cultivars by crossing them with wild or genetically remote cultivars. However, hybrids are rarely obtained from such distant crosses due to multitude of reproductive barriers. Interestingly, we found a fertile tetraploid progeny derived from anther culture of Asian rice cultivar, O. sativa, and African rice cultivar, O. glaberrima. Using genomic and phenotypic analyses, we now aim to clarify mechanisms involved in recovery of seed set in these plants. Furthermore, we hope to establish a novel remote cross breeding strategy in rice that overcomes reproductive barriers by introduction of polyploidization and haploidization.
For breeding of super rice varieties with useful new genes, it is important to promote allelic exchange in existing cultivars by crossing them with wild or genetically remote cultivars. However, hybrids are rarely obtained from such distant crosses due to multitude of reproductive barriers. Interestingly, we found a fertile tetraploid progeny derived from anther culture of Asian rice cultivar, O. sativa, and African rice cultivar, O. glaberrima. Using genomic and phenotypic analyses, we now aim to clarify mechanisms involved in recovery of seed set in these plants. Furthermore, we hope to establish a novel remote cross breeding strategy in rice that overcomes reproductive barriers by introduction of polyploidization and haploidization.
 |
|
|---|
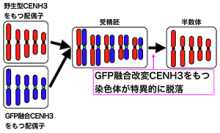
Design and synthesis of ‘chromosomes’ carrying new genome information
Nuclei that have very complex structures and various functions are the most important organelles in eukaryotic cells. Nuclear DNA are packed into chromosomes, enabling the accurate transmission of genetic information to daughter cells. Our research group is studying the molecular structures and functions of nuclei and chromosomes in plants. Our most recent goal is elucidate chromosome functional elements including centromeres. We are also interested in the relation between chromatin modifications and gene expression.
 |
|
|---|
Crop breeding boosted by genomics and bioinformatics
 Computers have revolutionized plant breeding, leading to the advent of ‘genomic breeding,’ which utilizes genome-wide information to model and predict phenotypic variation. Furthermore, recent advancements in DNA sequencing technology have generated vast amounts of genome and transcriptome data. However, the application of this data to practical breeding has been limited. Our research group aims to develop efficient methods to integrate various omics data for the exploration of agriculturally valuable genes and the production of new breeding materials.
Computers have revolutionized plant breeding, leading to the advent of ‘genomic breeding,’ which utilizes genome-wide information to model and predict phenotypic variation. Furthermore, recent advancements in DNA sequencing technology have generated vast amounts of genome and transcriptome data. However, the application of this data to practical breeding has been limited. Our research group aims to develop efficient methods to integrate various omics data for the exploration of agriculturally valuable genes and the production of new breeding materials.